Inclusion and Interpretation of Catgorical Predictors in Linear Regression Models
February 23, 2026
Motivation
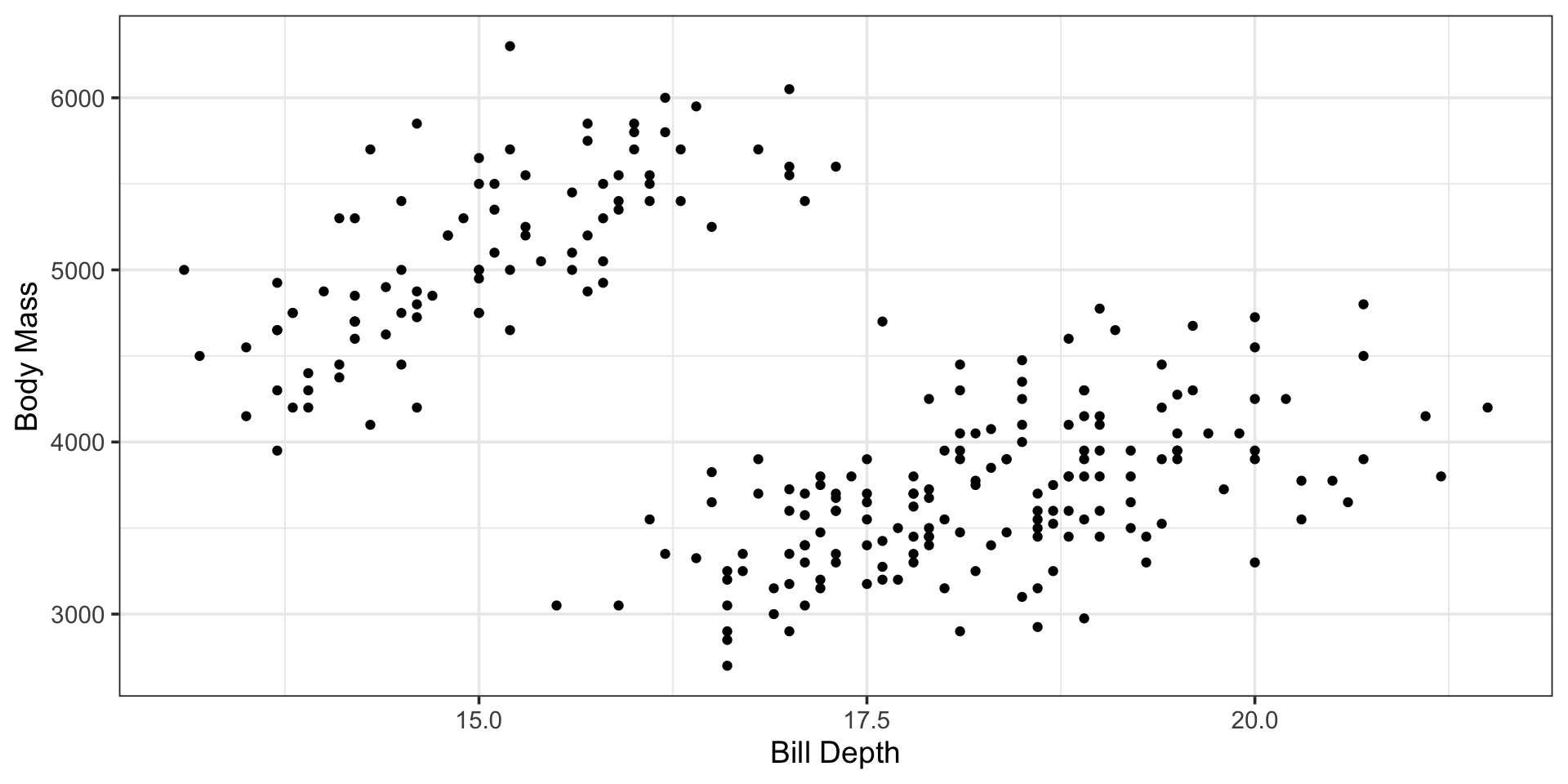
Motivation
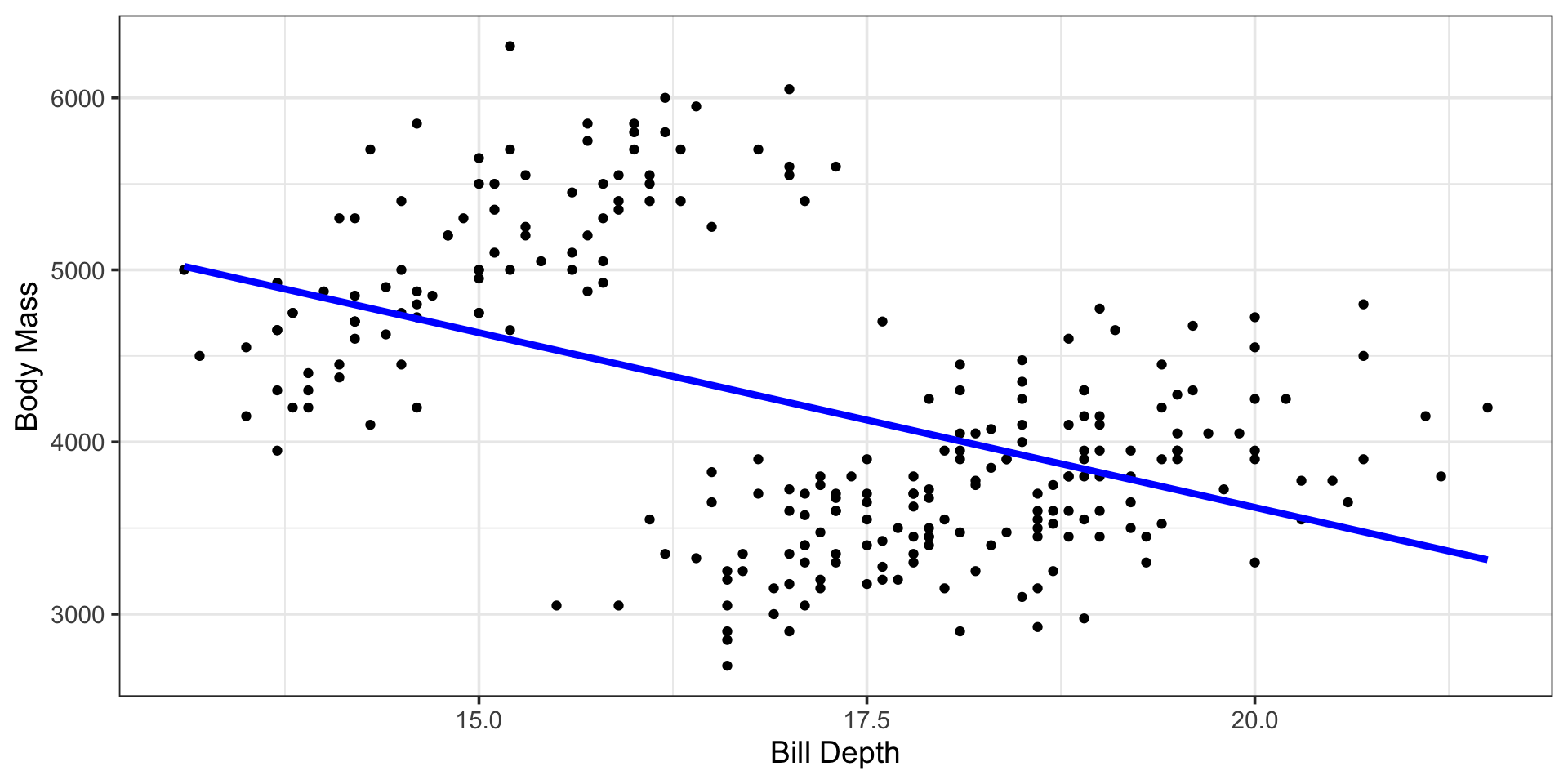
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.2260831 | 0.2230361 | 723.2657 | 74.2006 | 0 | 1 | -2047.691 | 4101.382 | 4112.018 | 132870759 | 254 | 256 |
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 7679.4255 | 406.12272 | 18.909126 | 0 |
| bill_depth_mm | -202.9711 | 23.56299 | -8.613977 | 0 |
Motivation
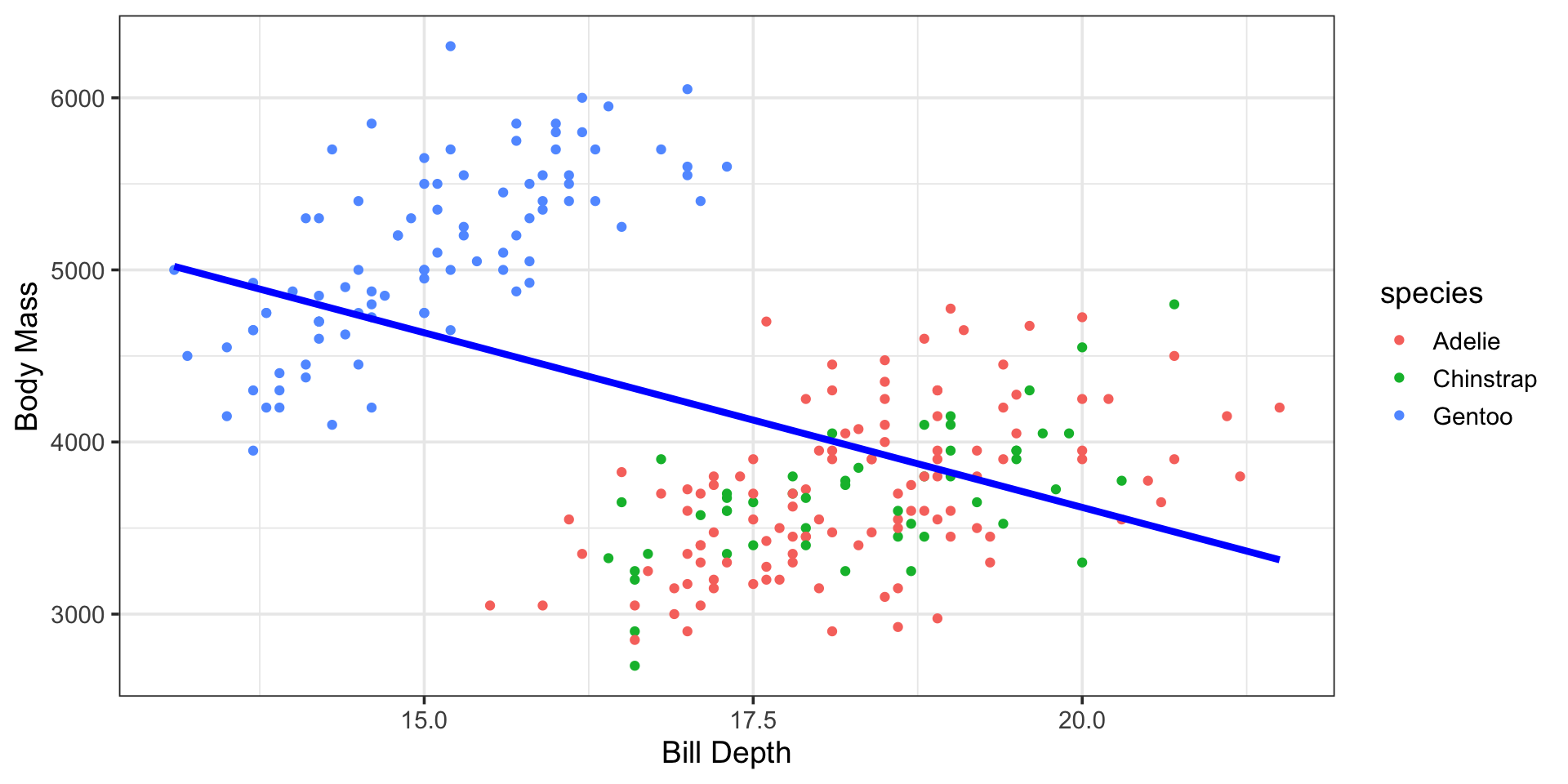
The Highlights
ANOVA as a linear regression model
Strategies for using categorical predictors in a model
- Ordinal scorings
- One-hot encodings versus dummy encodings
Feature engineering steps in recipes
Handling unknown or novel levels of a categorical variable
Fitting a model with categorical and numerical variables
Interpreting models with categorical predictors
Playing Along
As always, you are encouraged to implement the ideas and techniques discussed here with your own data. You should…
- Open RStudio and ensure that you are working within your
MAT300project space - Open the notebook which includes your models for Air BnB rentals
- Run all of the code in that notebook
You’ll add to that notebook here
ANalysis Of VAriance
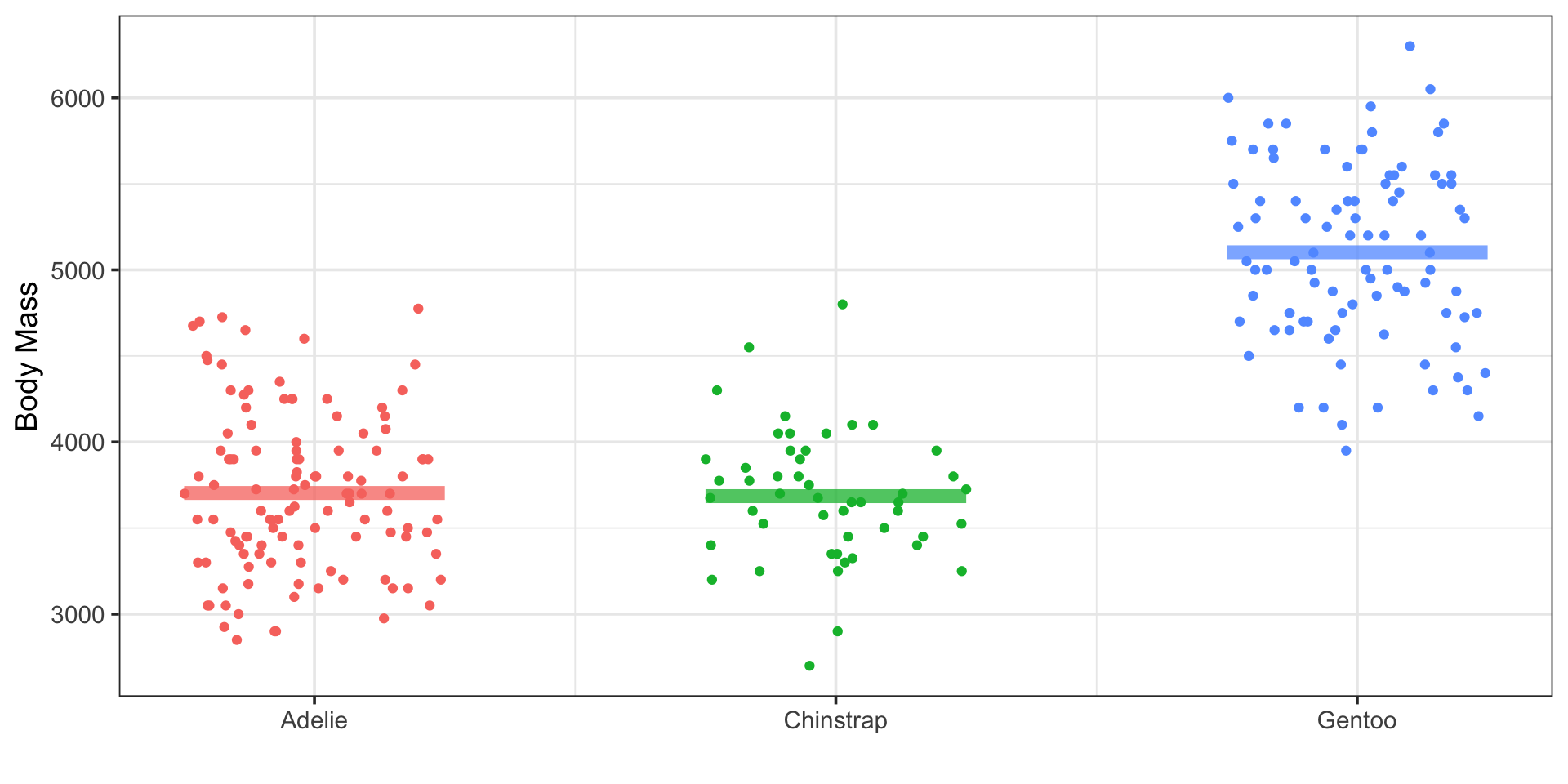
Does penguin body mass vary by species?
\[\begin{array}{lcl} H_0 & : & \mu_{\text{Adelie}} = \mu_{\text{Chinstrap}} = \mu_{\text{Gentoo}}\\ H_a & : & \text{At least one species has different average body mass}\end{array}\]
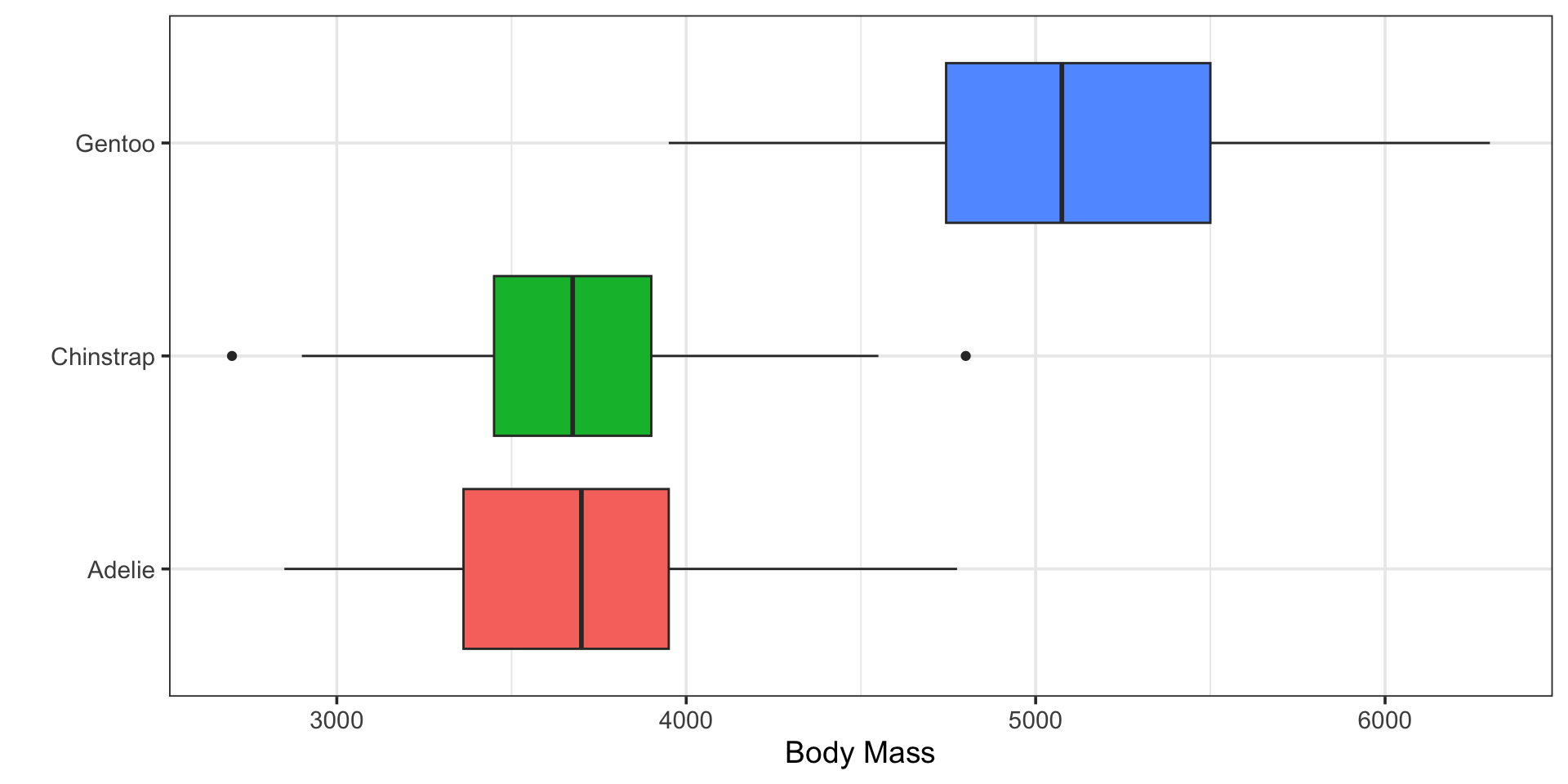
ANalysis Of VAriance
Does penguin body mass vary by species?
\[\begin{array}{lcl} H_0 & : & \mu_{\text{Adelie}} = \mu_{\text{Chinstrap}} = \mu_{\text{Gentoo}}\\ H_a & : & \text{At least one species has different average body mass}\end{array}\]
ANOVA Test:
| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| species | 2 | 116165670 | 58082835.2 | 264.6767 | 0 |
| Residuals | 253 | 55520401 | 219448.2 | NA | NA |
ANalysis Of VAriance as Linear Regression
Does penguin body mass vary by species?
\[\mathbb{E}\left[\text{body mass}\right] = \beta_0 + \beta_1\cdot\left(\text{???}\right)\]
Model-Based Test:
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.6766167 | 0.6740604 | 468.453 | 264.6767 | 0 | 2 | -1935.995 | 3879.99 | 3894.171 | 55520401 | 253 | 256 |
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 3703.94737 | 43.87464 | 84.4211365 | 0.0000000 |
| species_Chinstrap | -18.44737 | 79.46036 | -0.2321581 | 0.8166029 |
| species_Gentoo | 1398.22654 | 65.65281 | 21.2972846 | 0.0000000 |
ANalysis Of VAriance as Linear Regression
Does penguin body mass vary by species?
\[\mathbb{E}\left[\text{body mass}\right] = \beta_0 + \beta_1\cdot\left(\text{speciesChinstrap}\right) + \beta_2\cdot\left(\text{speciesGentoo}\right)\]
Model-Based Test:
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.6766167 | 0.6740604 | 468.453 | 264.6767 | 0 | 2 | -1935.995 | 3879.99 | 3894.171 | 55520401 | 253 | 256 |
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 3703.94737 | 43.87464 | 84.4211365 | 0.0000000 |
| species_Chinstrap | -18.44737 | 79.46036 | -0.2321581 | 0.8166029 |
| species_Gentoo | 1398.22654 | 65.65281 | 21.2972846 | 0.0000000 |
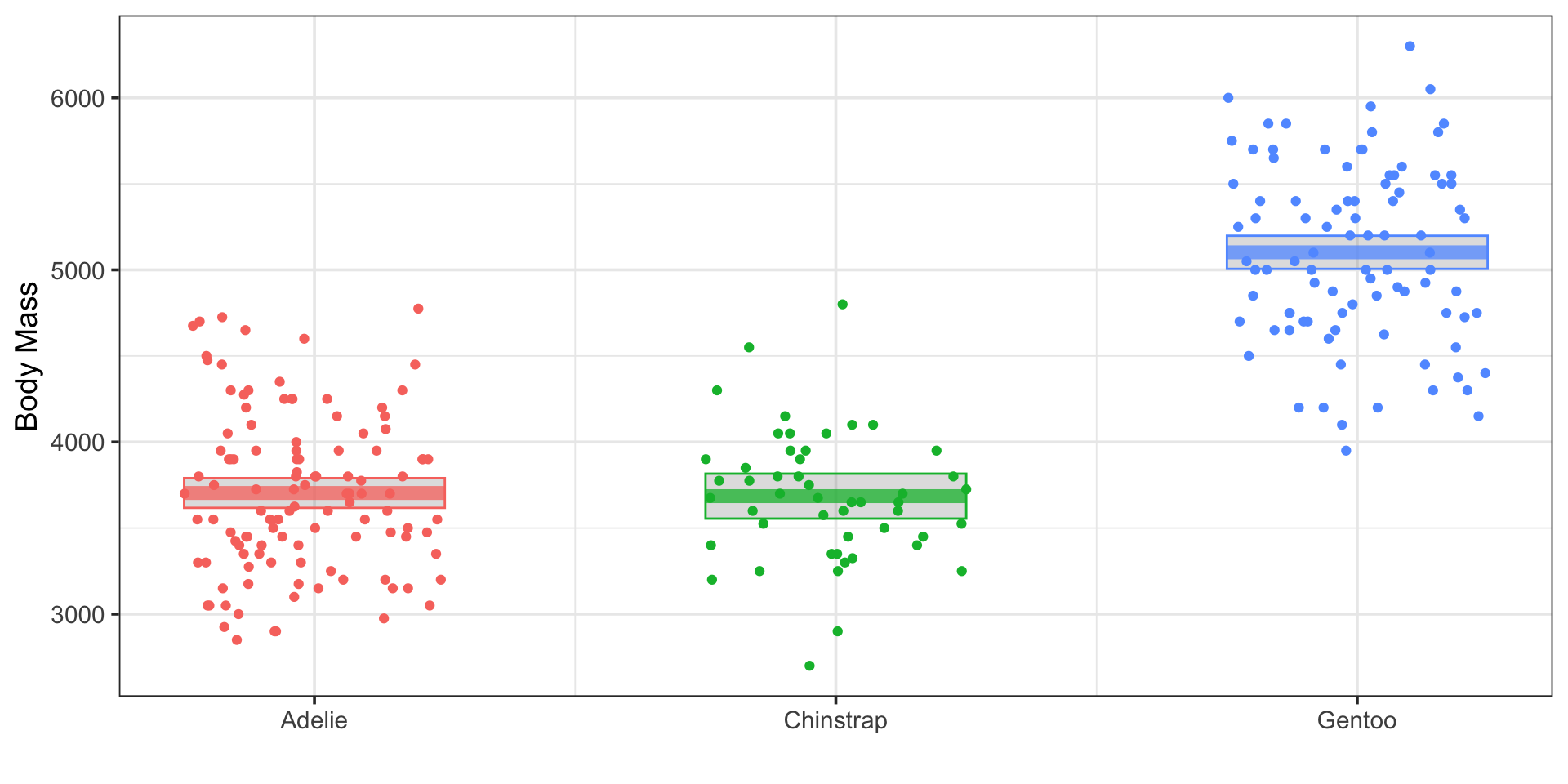
ANalysis Of VAriance as Linear Regression
Does penguin body mass vary by species?
\[\mathbb{E}\left[\text{body mass}\right] \approx 3703.95 - 18.45\cdot\left(\text{speciesChinstrap}\right) + 1298.23\cdot\left(\text{speciesGentoo}\right)\]
ANalysis Of VAriance as Linear Regression
Does penguin body mass vary by species?
\[\mathbb{E}\left[\text{body mass}\right] \approx 3703.95 - 18.45\cdot\left(\text{speciesChinstrap}\right) + 1298.23\cdot\left(\text{speciesGentoo}\right)\]
Categorical Predictors in Models
Linear regression models depend on multiplication and addition
These operations are not meaningful for categories (Adelie, Torgersen, red, etc.)
\[\mathbb{E}\left[\text{body mass}\right] = \beta_0 + \beta_1\cdot\left(\text{species}\right)\]
How would we evaluate this model for say, a Chinstrap penguin?
\[\begin{align} \mathbb{E}\left[\text{body mass}\right] &= \beta_0 + \beta_1\cdot\left(\text{Chinstrap}\right)\\ &= \text{???} \end{align}\]
We need some way to convert categories into numeric quantities so that our models can consume them
Strategies for Using Categorical Predictors (Scoring)
We can use an ordinal scoring method
| Species | Score |
|---|---|
| Adelie | 1 |
| Chinstrap | 2 |
| Gentoo | 3 |
Advantages:
- Only one \(\beta\)-coefficient required for inclusion of the categorical predictor
- Can accommodate/model rankings between levels
Drawbacks:
Imposes an ordering on categories
Enforces relationships between effect sizes
- For example, the effect of being Gentoo on body mass is \(3\times\) the effect of being Adelie
- The expected difference in body mass between Adelie and Chinstrap penguins is the same as the expected difference between Chinstrap and Gentoo penguins
Strategies for Using Categorical Predictors (Dummy Variables)
We can use dummy (or indicator) variables to encode the category
| species | speciesAdelie | speciesChinstrap | speciesGentoo |
|---|---|---|---|
| Gentoo | 0 | 0 | 1 |
| Adelie | 1 | 0 | 0 |
| Gentoo | 0 | 0 | 1 |
| Chinstrap | 0 | 1 | 0 |
| Adelie | 1 | 0 | 0 |
| Chinstrap | 0 | 1 | 0 |
Strategies for Using Categorical Predictors (Dummy Variables)
We can use dummy (or indicator) variables to encode the category
So we don’t need the species variable any longer
| speciesAdelie | speciesChinstrap | speciesGentoo |
|---|---|---|
| 0 | 0 | 1 |
| 1 | 0 | 0 |
| 0 | 0 | 1 |
| 0 | 1 | 0 |
| 1 | 0 | 0 |
| 0 | 1 | 0 |
Strategies for Using Categorical Predictors (Dummy Variables)
We can use dummy (or indicator) variables to encode the category
So we don’t need the species variable any longer
And we don’t need every indicator column, either
| speciesChinstrap | speciesGentoo |
|---|---|
| 0 | 1 |
| 0 | 0 |
| 0 | 1 |
| 1 | 0 |
| 0 | 0 |
| 1 | 0 |
Strategies for Using Categorical Predictors (Dummy Variables)
We can use dummy (or indicator) variables to encode the category
So we don’t need the species variable any longer
And we don’t need every indicator column, either
| species | speciesChinstrap | speciesGentoo |
|---|---|---|
| Gentoo | 0 | 1 |
| Adelie | 0 | 0 |
| Gentoo | 0 | 1 |
| Chinstrap | 1 | 0 |
| Adelie | 0 | 0 |
| Chinstrap | 1 | 0 |
Advantage: Can model variable effect sizes between levels
Disadvantage: Requires more \(\beta\)-coefficients (one less than the number of unique levels)
Strategies for Using Categorical Predictors
\(\bigstar\) Let’s try it! \(\bigstar\)
- Choose one categorical predictor of price for a rental in your dataset
- How many levels of your categorical variable are there?
- Is a scoring method meaningful for the levels of your variable? Why or why not?
- What would a dummy-encoding of your categorical variable look like? How many indicator columns would be introduced?
Back to Our Model from Earlier
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 3703.94737 | 43.87464 | 84.4211365 | 0.0000000 |
| species_Chinstrap | -18.44737 | 79.46036 | -0.2321581 | 0.8166029 |
| species_Gentoo | 1398.22654 | 65.65281 | 21.2972846 | 0.0000000 |
\[\mathbb{E}\left[\text{body mass}\right] \approx 3703.95 - 18.45\cdot\left(\text{speciesChinstrap}\right) + 1398.23\cdot\left(\text{speciesGentoo}\right)\]
Prediction for Adelie: \(3703.95 - 18.45\cdot\left(0\right) + 1398.23\cdot\left(0\right) \approx 3703.95\text{g}\)
Prediction for Chinstrap: \(3703.95 - 18.45\cdot\left(1\right) + 1398.23\cdot\left(0\right) \approx 3685.5\text{g}\)
Prediction for Gentoo: \(3703.95 - 18.45\cdot\left(0\right) + 1398.23\cdot\left(1\right) \approx 5102.18\text{g}\)
Note: The interpretations for these \(\beta\)-coefficients on the species dummy variables are not as slopes. They are direct adjustments to the expected body mass, accounting for differences in species. The intercept in this model is the predicted response for the base level.
Including Categorical Predictors with {tidymodels}
The act of transforming a categorical column into either ordinal scores or dummy variables is a feature-engineering step.
We’ll encounter more feature engineering steps throughout our course. In the {tidymodels} framework, feature engineering steps are carried out as steps appended to our recipe().
To build the linear regression version of our ANOVA test, I used the following to set up the model:
I then used our usual code to construct the workflow and fit the model to our training data.
Including Categorical Predictors with {tidymodels}
\(\bigstar\) Let’s try it! \(\bigstar\)
Add a new section to your notebook on the use of categorical predictors in models
Set up a linear regression model which uses your chosen categorical variable as the sole predictor of rental price
- Create a model specification
- Create a recipe and use the
step_dummy()feature-engineering step in order to convert your categorical column into a collection of indicator columns - Package your specification and recipe together into a workflow
- Fit the workflow to the training data
Examine the global and term-based model metrics
Write down the equation of the model
Determine the resulting model for several of the levels of your categorical variable
Something More Interesting
Let’s build a model that better corresponds to the plot we began this discussion with…
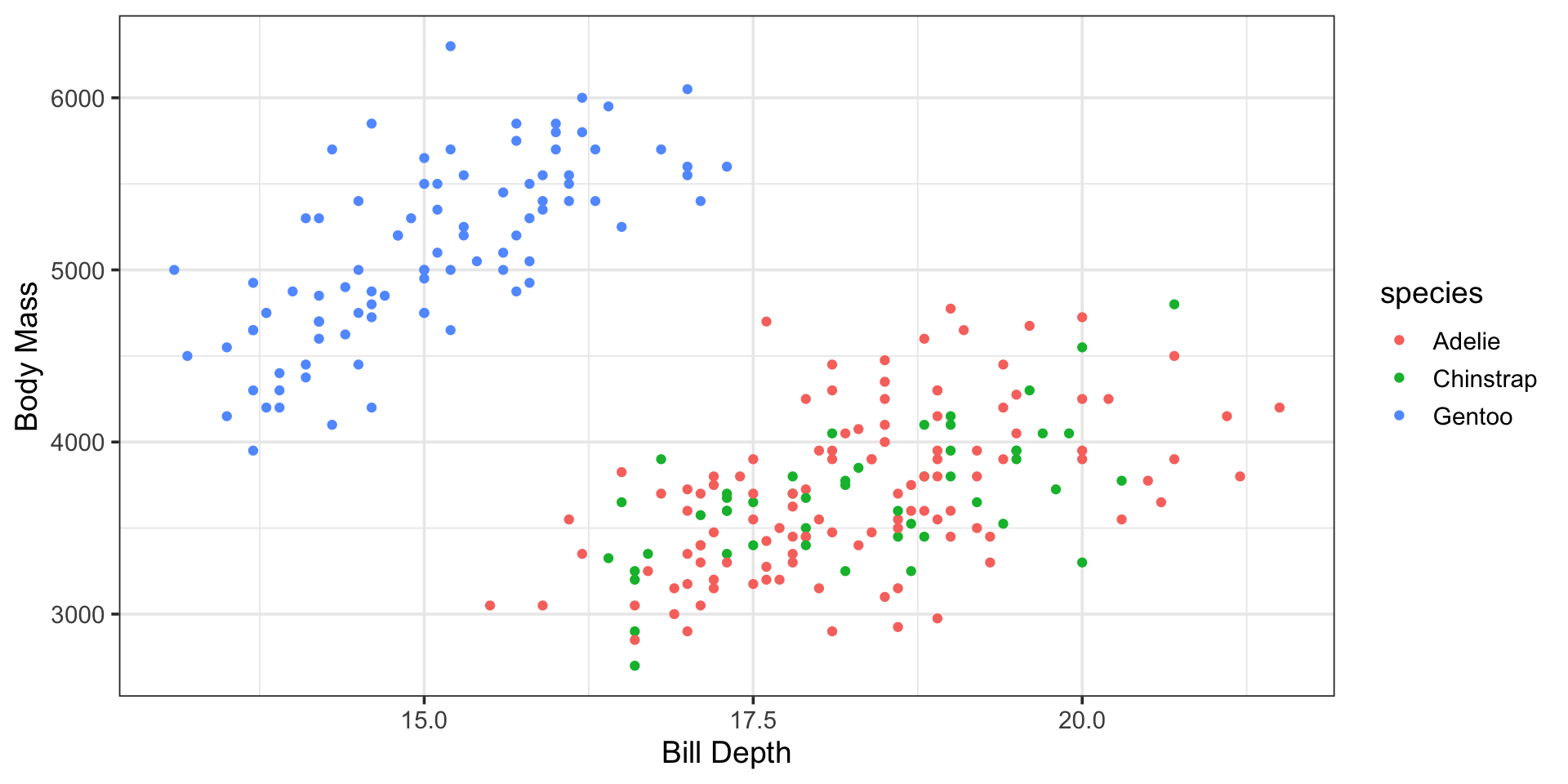
Can we accomodate both bill_depth_mm (a numerical variable) and species (a categorical variable) into a single model?
Bill Depth and Species as Predictors of Body Mass
Now our recipe, with the feature engineering step included
Next, we package both the model and recipe together into a workflow
Important Note! Common concerns and remedies when including categorical predictors…
step_unknown(cat_var_name)to fill in missing values withunknownstep_novel(cat_var_name)to reassign new classes to anovelclassstep_other(cat_var_name)to combine rare classes into anotherclass
Models with Numerical and Categorical Predictors
\(\bigstar\) Let’s try it! \(\bigstar\)
- Create an instance of a modeling workflow which uses at least one numerical predictor and at least one categorical predictor
- Fit your workflow to your training data
Assessing Our New Model
Global Test for Model Utility:
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.7928019 | 0.7903352 | 375.7164 | 321.4091 | 0 | 3 | -1879.014 | 3768.028 | 3785.754 | 35573029 | 252 | 256 |
Individual Term-Based Tests:
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|
| (Intercept) | -912.90624 | 389.97673 | -2.3409249 | 0.0200163 | -1680.9351 | -144.8773 |
| bill_depth_mm | 252.33546 | 21.22734 | 11.8872854 | 0.0000000 | 210.5299 | 294.1411 |
| species_Chinstrap | -23.37012 | 63.73145 | -0.3666969 | 0.7141529 | -148.8843 | 102.1440 |
| species_Gentoo | 2220.99703 | 86.96707 | 25.5383665 | 0.0000000 | 2049.7221 | 2392.2719 |
Assessing Our New Model
Individual Term-Based Tests:
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|
| (Intercept) | -912.90624 | 389.97673 | -2.3409249 | 0.0200163 | -1680.9351 | -144.8773 |
| bill_depth_mm | 252.33546 | 21.22734 | 11.8872854 | 0.0000000 | 210.5299 | 294.1411 |
| species_Chinstrap | -23.37012 | 63.73145 | -0.3666969 | 0.7141529 | -148.8843 | 102.1440 |
| species_Gentoo | 2220.99703 | 86.96707 | 25.5383665 | 0.0000000 | 2049.7221 | 2392.2719 |
Note: Controlling for differences in bill depth, the average body mass for Gentoo and Adelie penguins is statistically discernible, but the body mass of Adelie and Chinstrap penguins is not.
\[\begin{align} \mathbb{E}\left[\text{body mass}\right] \approx -912.91 ~+ &~252.34\cdot\left(\text{bill depth}\right) - 23.37\cdot\left(\text{speciesChinstrap}\right) +\\ &~2221.0\cdot\left(\text{speciesGentoo}\right)\end{align}\]
An Improvement Over the Species-Only Model?
Species-Only Model:
| metric | value |
|---|---|
| r.squared | 0.6766167 |
| adj.r.squared | 0.6740604 |
| sigma | 468.4530146 |
| statistic | 264.6767122 |
| p.value | 0.0000000 |
| df | 2.0000000 |
| logLik | -1935.9949729 |
| AIC | 3879.9899458 |
| BIC | 3894.1706556 |
| deviance | 55520401.4016019 |
| df.residual | 253.0000000 |
| nobs | 256.0000000 |
Species and Bill Depth Model:
| metric | value |
|---|---|
| r.squared | 0.7928019 |
| adj.r.squared | 0.7903352 |
| sigma | 375.7164021 |
| statistic | 321.4091063 |
| p.value | 0.0000000 |
| df | 3.0000000 |
| logLik | -1879.0141347 |
| AIC | 3768.0282694 |
| BIC | 3785.7541567 |
| deviance | 35573029.3286923 |
| df.residual | 252.0000000 |
| nobs | 256.0000000 |
Assessing and Comparing Models
\(\bigstar\) Let’s try it! \(\bigstar\)
- Obtain and interpret your global and term-based model metrics
- Compare your model’s training metrics to those of your previous models
- Obtain performance metrics for this new model using the test data. How does this model perform on unknown test observations compared to your previous models?
Visualizing Model Coefficients
| term | estimate | std.error | p.value | conf.low | conf.high |
|---|---|---|---|---|---|
| (Intercept) | -912.90624 | 389.97673 | 0.0200163 | -1680.9351 | -144.8773 |
| bill_depth_mm | 252.33546 | 21.22734 | 0.0000000 | 210.5299 | 294.1411 |
| species_Chinstrap | -23.37012 | 63.73145 | 0.7141529 | -148.8843 | 102.1440 |
| species_Gentoo | 2220.99703 | 86.96707 | 0.0000000 | 2049.7221 | 2392.2719 |
Code
lr_sbl_fit %>%
extract_fit_engine() %>%
tidy(conf.int = TRUE) %>%
ggplot() +
geom_errorbarh(aes(xmin = conf.low, xmax = conf.high, y = term, color = term),
lwd = 1.25) +
geom_point(aes(x = estimate, y = term, color = term),
size = 4) +
geom_vline(xintercept = 0, linetype = "dashed", lwd = 1.25) +
labs(title = "Model Coefficients",
x = "Coefficient",
y = "") +
theme(legend.position = "None")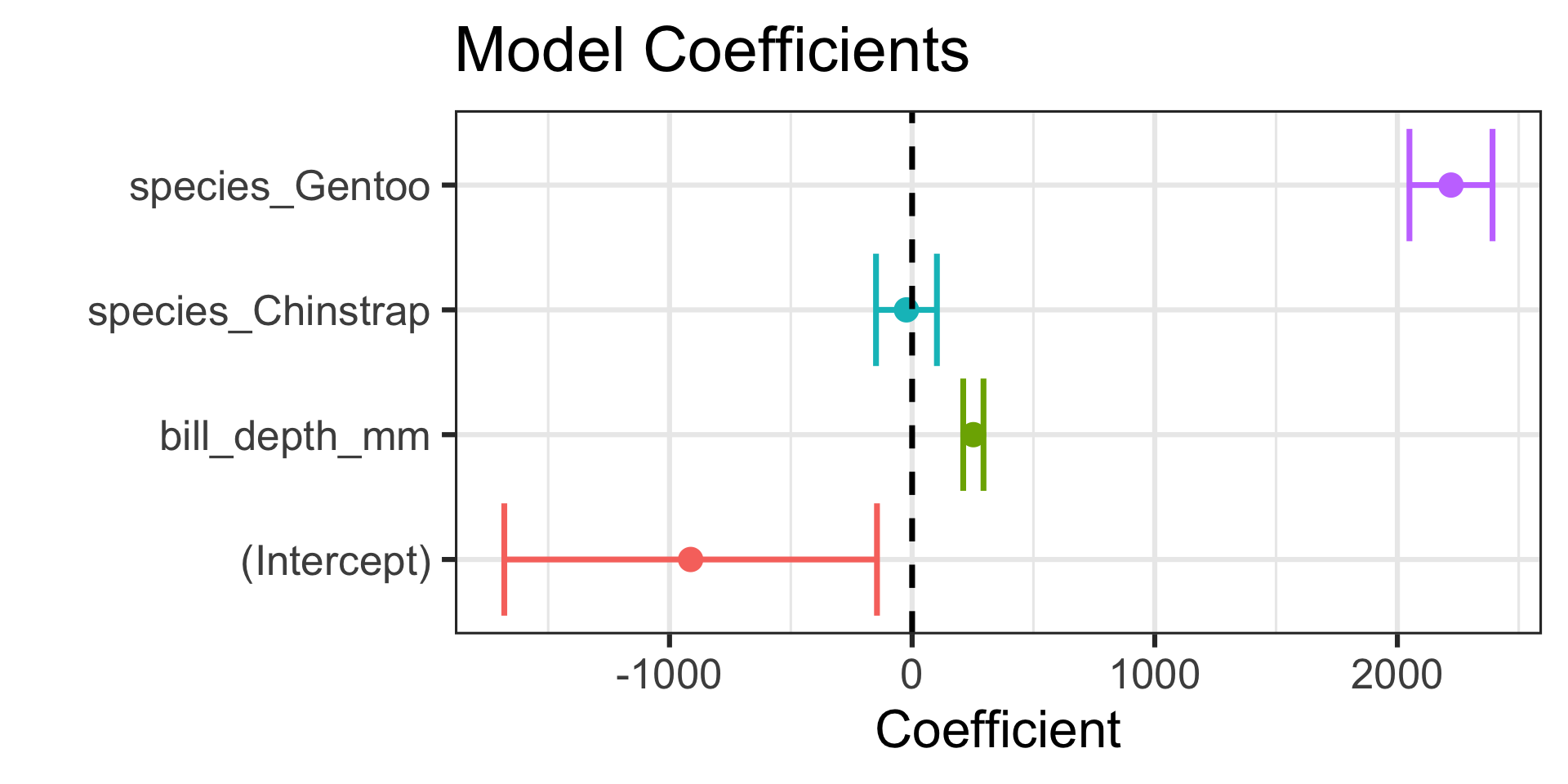
Visualizing Coefficients
\(\bigstar\) Let’s try it! \(\bigstar\)
- Try to construct a plot which allows you to visualize your model coefficients and their plausible ranges.
- What does the plot tell you about each model term?
Interpreting this Model
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -912.90624 | 389.97673 | -2.3409249 | 0.0200163 |
| bill_depth_mm | 252.33546 | 21.22734 | 11.8872854 | 0.0000000 |
| species_Chinstrap | -23.37012 | 63.73145 | -0.3666969 | 0.7141529 |
| species_Gentoo | 2220.99703 | 86.96707 | 25.5383665 | 0.0000000 |
\[\begin{align} \mathbb{E}\left[\text{body mass}\right] \approx -912.91 ~+ &~252.34\cdot\left(\text{bill depth}\right) - 23.37\cdot\left(\text{speciesChinstrap}\right) +\\ &~2221.0\cdot\left(\text{speciesGentoo}\right)\end{align}\]
(Intercept) Interpretation of this intercept is not meaningful because it would correspond to a penguin with a bill depth of 0mm.
(Bill Depth) Controlling for differences in species, we expect a 1mm increase in bill depth to be associated with a 252.34g increase in body mass, on average.
(Chinstrap) Given a Chinstrap and Adelie penguin with the same bill depth, we estimate the Chinstrap to have a lower body mass by about 23.37g, but this is not a statistically discernible difference.
(Gentoo) Given a Gentoo and Adelie penguin with the same bill depth, we expect the Gentoo to have greater mass by about 2221.0g, on average.
- Note: Some comparisons are misleading since Adelie bill depths rarely overlap with Gentoos’
Interpreting the Model
\(\bigstar\) Let’s try it! \(\bigstar\)
- If you haven’t done so already, write down your estimated model form.
- Interpret the intercept (if appropriate) and the estimated effect of each predictor on the response.
Making Predictions with this Model
new_data <- crossing(
bill_depth_mm = seq(13.1, 21.5, length.out = 250),
species = c("Adelie", "Chinstrap", "Gentoo")
)
new_data <- lr_sbl_fit %>%
augment(new_data) %>%
bind_cols(
lr_sbl_fit %>%
predict(new_data, type = "conf_int") %>%
rename(
.conf_lower = .pred_lower,
.conf_upper = .pred_upper
)
) %>%
bind_cols(
lr_sbl_fit %>%
predict(new_data, type = "pred_int")
)Making Predictions with this Model
ggplot() +
geom_ribbon(data = new_data,
aes(x = bill_depth_mm, ymin = .pred_lower, ymax = .pred_upper),
alpha = 0.25) +
geom_ribbon(data = new_data,
aes(x = bill_depth_mm, ymin = .conf_lower, ymax = .conf_upper),
alpha = 0.5) +
geom_line(data = new_data,
aes(x = bill_depth_mm, y = .pred, color = species),
lwd = 1.25) +
geom_point(data = penguins_train,
aes(x = bill_depth_mm, y = body_mass_g, color = species),
alpha = 0.5) +
labs(
x = "Bill Depth",
y = "Body Mass"
) +
facet_wrap(~species, ncol = 3) +
theme(legend.position = "None")Making Predictions with this Model
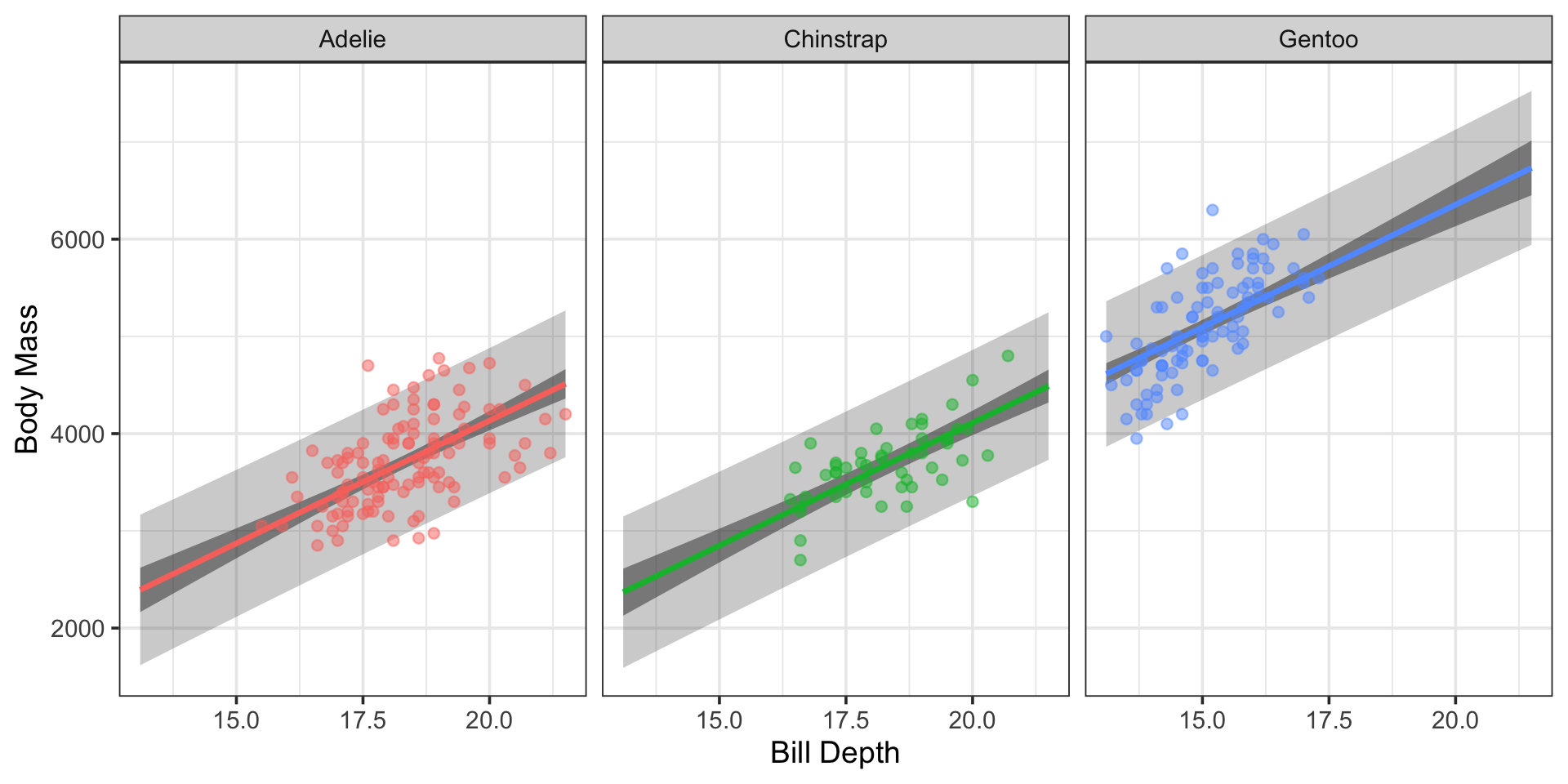
Note: Including the categorical predictor has resulted in our model taking the form of these separate, parallel lines.
Making and Visualizing Model Predictions
\(\bigstar\) Let’s try it! \(\bigstar\)
Create a counterfactual set of new observations using
crossing()Use your model to make predictions for those new observations
- Include confidence intervals and prediction intervals as well
Create a graphical representation of your model’s predictions, including confidence- and prediction-bands
Additional Feature-Engineering Steps for Categorical Predictors
- If we expect to encounter missing values, we can use
step_unknown()to create a level for unknown values or we can usestep_impute_mode()to fill in missing levels with the most frequently observed level - If we expect to encounter new levels, unknown to the model at training time, we can use
step_novel() - If we expect to have levels which are infrequently observed, we can group those levels together into an “
other” level withstep_other()
Summary
We can include categorical predictors in a model to differentiate predictions across the levels of that variable
Including categorical predictors in a model requires the use of an encoding scheme in order to convert levels to numerical values
Scoring assigns a distinct number to each category
- Implement it by adding
step_ordinalscore()as a feature engineering step in a recipe
- Implement it by adding
Dummy encodings replace a categorical predictor with a collection of binary (0/1) variables indicating the levels of the categorical predictor
- We can use the dummy variable technique by adding
step_dummy()as a feature engineering step in a recipe
- We can use the dummy variable technique by adding
In the case of rare levels, we may want to also include
step_other(),step_unknown(), orstep_novel()as recipe steps as well – order of application matters!
A linear regression model including a single categorical variable as the sole predictor is equivalent to ANOVA
For a linear regression model with at least one numerical predictor, the result of the inclusion of a categorical variable in the model is a shift in intercept (a vertical shift) for each of the levels of the categorical predictor
Next Time…
Model-Building, Assessment, and Interpretation Workshop